The breeding data TRIAD: Phenotyping, Pedigree, Genotyping
Dr. Salvador A. Gezan
16 March 2022
The analysis of breeding data can be quite complex, requiring management of large databases that come from different types of studies, and that often have a messy structure. This is to be expected as breeding programs are long-term enterprises spanning over several generations, using technologies that evolve over time, and with changing leadership and funding. Therefore, breeding programs require careful data management and statistical tools and expertise to extract the relevant information for us to select outstanding genotypes that will form the next generation of our commercial population.
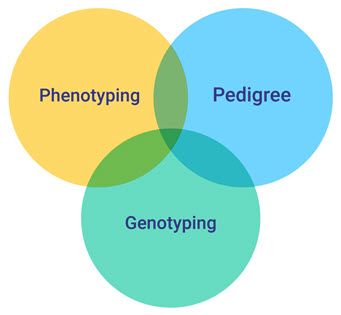
One easy way to visualize this complexity is by thinking of the breeding triad, as shown in the figure below. This triad is formed by phenotyping, the pedigree and genotyping. In the following sections we will describe them, and some of the challenges and issues that are associated with them.
The term phenotype refers to the observable physical properties of an organism; these include the organism's appearance, development, and behaviour. Some important aspects are:
- The evaluation of the performance of an individual, or group of individuals, will ultimately require that some physical measurements are available
- The ultimate goal of phenotypic data is to fit models to predict performance as breeding or total genetic values, on the same or related individuals
A pedigree is a genetic representation of a family tree that describes the inheritance though several generations. This is crucial for most breeding programs, given that pedigree allows us to estimate breeding values by correlating genetic entities (e.g., full-sibs). Some important aspects are:
- It is very useful for many tasks to keep track of historical and current relatedness and inbreeding
- Pedigree information also assists with operational decisions and planning/executing crossing schemes for future breeding and commercial populations
- There is value-added to pedigree once it is combined with genomic information, particularly if we have a large amount of historical data
An organism's phenotype is determined by its genotype, which is the set of genes the organism carries. These genes, which are encoded in the DNA, are then used to make proteins and RNA molecules, which in turn determine the individual’s traits.
- This genotype is valuable information to assess our breeding population (genetic diversity, population structure, etc.)
- All modern predictive genomic tools use this information (particularly SNPs), including:
- Genomic prediction (estimation of breeding values)
- Genome-wide association (identification of specific SNP markers)
- Parentage reconstruction
- Optimization of crossing and mating schemes
Challenges in breeding data management and integration
As indicated above, all three elements of the breeding triad have a role to play in the success of a breeding program. They require a dedicated investment both for producing the above information, and also for managing and curating the data they generate. And this is not an easy task! In the last decades, with the emergence of genomic tools, this has become even more complex. This new scenario is creating many challenges, both computational and analytical, in our workflows. A few relevant points are mentioned below.
- The greatest problem is combining all pieces into a single system that integrates them well. We need additional checks to verify the data, requiring new programming routines and procedures in place
- Information grows steadily creating challenges with database structure, storing, accessing, and statistical processing. Moreover, it is likely our personal computers are not be able to handle this data efficiently, and hence, we need more RAM memory and possibly a cloud system to manage this growing data
- Statistical analyses (and breeding models) are becoming more complex, requiring additional computational resources. Again, we need to make sure our infrastructure can handle this. Nevertheless, more data allows us to explore and develop different models: like those used in machine learning!
- Storing, managing and curating data is an additional expense to our breeding program, on top of phenotyping and genotyping, that needs to have a secure and adequately funded budget
Investing in breeding data: A strategic perspective
Let’s not forget that as we accumulate more and more breeding information this data will become (and often is at the present) one of our most important investments. We need to keep in mind that this data will have more (known and unknown) uses in the future. A few of the known uses include:
- Construction (i.e., training) of predictive models (they all require data!)
- Use of modern modelling tools (e.g., machine learning, AI)
- Exploring and obtaining the benefits of high-throughput phenotyping (HTP)
- Keeping better track of genomic changes/progress in a population
- Maintaining a breeding strategy that will provide us with genetic gains in the short- and long-term
All data, but particularly genomic information, will become an important economic and strategic asset!
Building a sustainable analytical and database system
In summary, let’s keep in mind this triad. We need to have a plan to maintain and manage it before we move into using complex, but more appropriate, statistical tools to exploit it. For this reason, it is important that for any breeding program we construct or design an analytical and database system that works with us from the early stages. And keep in mind that this is not going to become any easier as we continue accumulating more data!
About the author
Salvador Gezan is a statistician/quantitative geneticist with more than 20 years’ experience in breeding, statistical analysis and genetic improvement consulting. He currently works as a Statistical Consultant at VSN International, UK. Dr. Gezan started his career at Rothamsted Research as a biometrician, where he worked with Genstat and ASReml statistical software. Over the last 15 years he has taught ASReml workshops for companies and university researchers around the world.
Dr. Gezan has worked on agronomy, aquaculture, forestry, entomology, medical, biological modelling, and with many commercial breeding programs, applying traditional and molecular statistical tools. His research has led to more than 100 peer reviewed publications, and he is one of the co-authors of the textbook “Statistical Methods in Biology: Design and Analysis of Experiments and Regression”.
Popular
Related Reads
