Uncovering disease control strategies: A Genstat case study on Maedi-Visna (MV) epidemiology in sheep
Uncovering disease control strategies: A Genstat case study on Maedi-Visna (MV) epidemiology in sheep
This case study focuses on the collaborative research of Andrew Illius, Emeritus Professor of Animal Ecology at the University of Edinburgh, and Nick Savill, Senior Lecturer in the School of Biological Sciences at the University of Edinburgh. They have been investigating the epidemiology and control of maedi-visna (MV), a debilitating lentivirus disease that infects sheep, goats, and wild ruminants causing inflammatory lesions to build up in the lungs, mammary gland, brain and the synovial membrane of joints. MV is a significant concern for farmers due to its rapid and silent spread within flocks, leading to welfare and productivity issues. By the time clinical signs are evident, the disease is well-established, with perhaps 30-70% of the flock infected. Through the use of Genstat, Andrew and Nick analysed long-term unbalanced experimental data, revealing crucial insights into disease control strategies.
Research objective
For a farmer, the disease raises crucial questions: How quickly will symptoms develop, and will welfare and productivity suffer? How soon will it spread through the rest of the flock? Is living with the disease more expensive than trying to eradicate it? The primary aim of this research was to find ways to control MV by analysing data from long-term experiments. MV is caused by a small ruminant lentivirus (SRLV). However, despite a substantial amount of research on the pathology, immunology, and molecular biology of SRLVs, the team were surprised to find there was a lack of quantitative analysis on SRLV epidemiology.
Modelling MV transmission
The research team leveraged a mathematical model developed by Nick Savill based on infectious disease epidemiology, and the known and unknown aspects of SRLV biology. This model estimated the probability of seroconversion, shedding light on the disease's transmission patterns. The study was the first to estimate transmission parameters, revealing that MV is strongly associated with housing, and transmission is negligible among sheep kept at grass. This finding presented an opportunity for disease control exploiting the fact that transmission of the virus is too slow between grazing sheep to sustain the disease.
Understanding disease spread
Further analysis focused on understanding infectiousness changes during an individual's infection. The team examined whether ewes with long-standing infections posed the greatest risk to uninfected sheep, and also explored management strategies to slow disease spread. Genstat was instrumental to this analysis.
Quantifying costs with Genstat
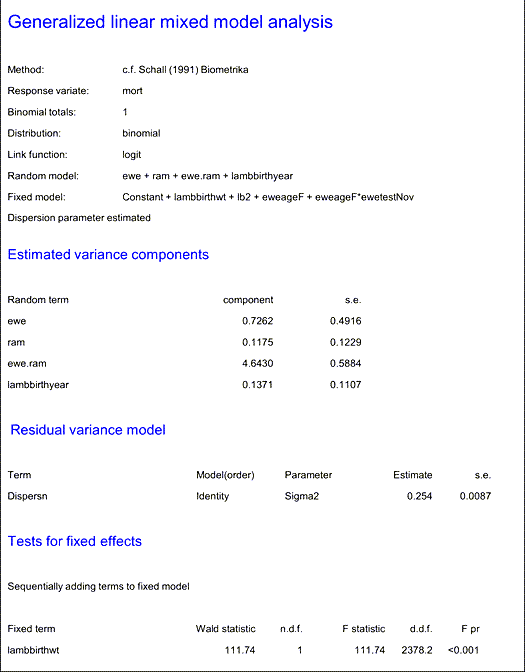
To assess the costs of living with MV, a large dataset from Lublin University of Life Sciences was analysed. This dataset included lambing records from about 800 ewes of different breeds, with over 300 ewes present each year. Given the complex nature of the data (unbalanced design, multiple observations from individual ewes and rams over several years, and different breeds) the team utilised Genstat to analyse it. The team employed mixed models to distinguish random and fixed effects, making use of the latest release of Genstat, which included Akaike’s Information Coefficient (AIC) for model selection. The analysis addressed pre-weaning lamb mortality and showed that the probability of mortality was strongly affected by lamb birthweight and the SRLV status of the ewe. Lamb growth rate was also shown to be negatively related to the time since the dam had become infected.
Genstat's role in the study
Andrew Illius, with decades of experience using Genstat, praised the software for its crucial role in agricultural and ecological research.
“The software's evolution from a Fortran-based system to a menu-driven environment on Windows significantly sped up analysis processes. Genstat's ability to handle unbalanced data and mixed models proved essential in this study. It’s a fantastic product, and a pleasure to use.”
Advancing scientific knowledge through Genstat
The research provides evidence-based insights for controlling MV, suggesting targeted disease control measures and social distancing strategies for sheep. This Genstat case study demonstrates the software's versatility in analysing complex data and its importance in advancing scientific knowledge.
By utilising Genstat, the collaborative research of Andrew Illius and Nick Savill contributes to understanding Maedi-Visna epidemiology, potentially leading to more effective disease control measures for the benefit of farmers and livestock alike.
Example output from Genstat